Published in J. Phys. Chem. Lett., 2014
Abstract
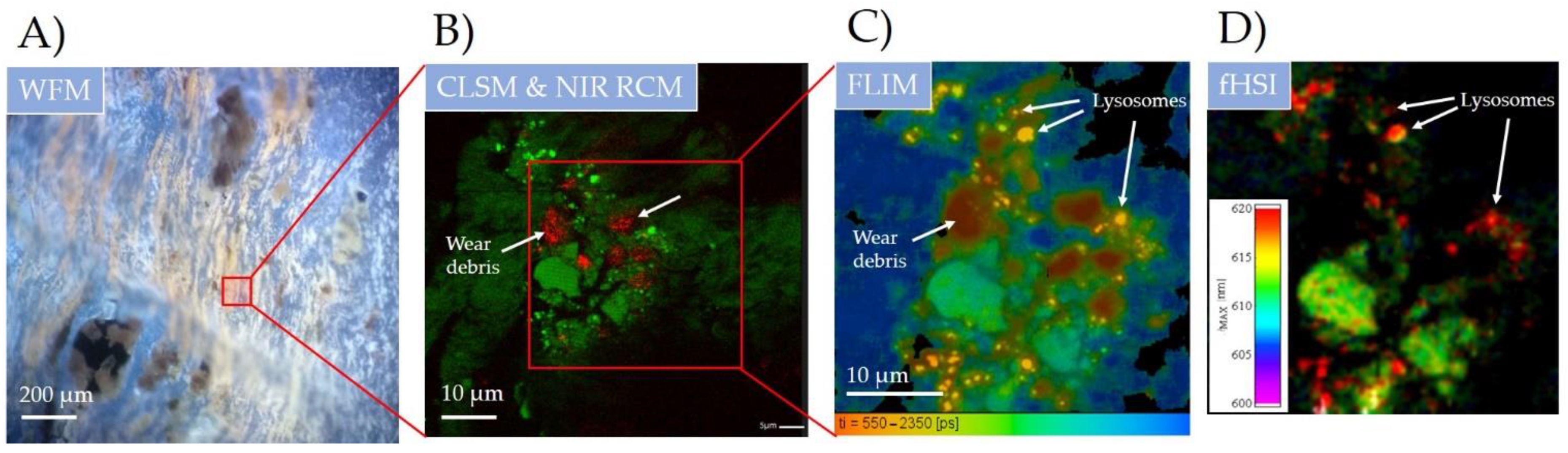
Conformational entropy (SΩ) has long been used to theoretically characterize the dynamics of proteins, DNA, and other polymers. Though recent advances enabled its calculation also from simulations and nuclear magnetic resonance (NMR) relaxation experiments, correlated molecular motion has hitherto greatly hindered both numerical and experimental determination, requiring demanding empirical and computational calibrations. Herein, we show that these motional correlations can be estimated directly from the temperature-dependent SΩ series that reveal effective persistence lengths of the polymers, which we demonstrate by measuring SΩ of amphiphilic molecules in model lipid systems by spin-labeling electron paramagnetic resonance (EPR) spectroscopy. We validate our correlation-corrected SΩ meter against the basic biophysical interactions underlying biomembrane formation and stability, against the changes in enthalpy and diffusion coefficients upon phase transitions, and against the energetics of fatty acid dissociation. As the method can be directly applied to conformational analysis of proteins and other polymers, as well as adapted to NMR or polarized fluorescence techniques, we believe that the approach can greatly enrich the scope of experimentally available statistical thermodynamics, offering new physical insights into the behavior of biomolecules.
Download here